Unit 9: Graphs
1 Graph Basics
Graphs aren’t new to us. Remember that a graph is a set of vertices, and the edges that connect them. We remember this because we’ve been dealing with trees, and trees, as we learned, are just a special form of graph.
Graphs are a spectacularly useful mathematical construct, and a lot of seemingly unrelated problems can be viewed as graph problems. This is a useful thing to have, because if somebody can solve a general graph problem, then they’ve solved every application that somebody can get to look like a graph problem.
What are some examples of graphs? Here’s the DC Metro system:
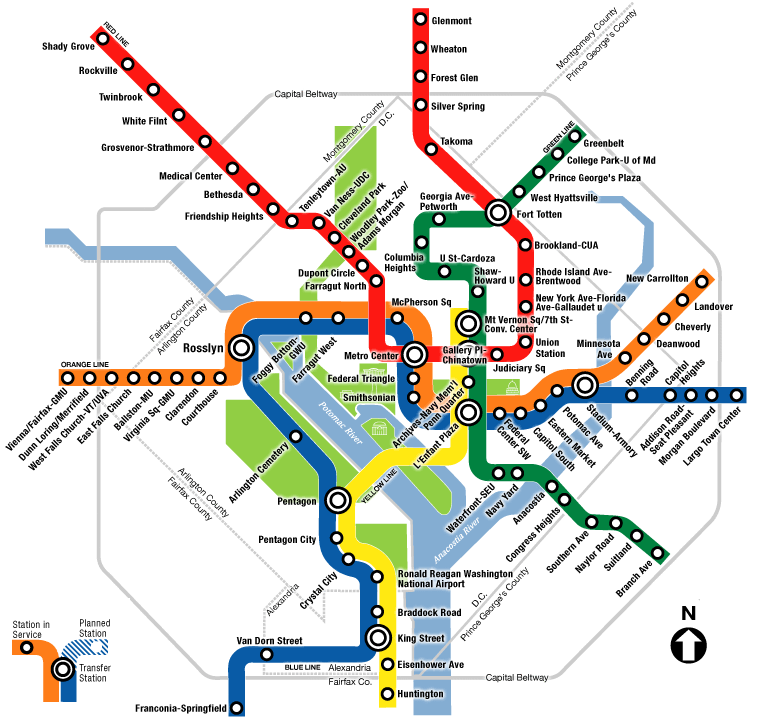
The map of the Metro is a graph where Metro stops are the vertices, and the routes the trains take are the edges.
We could also draw the friendships in a social network as a graph:
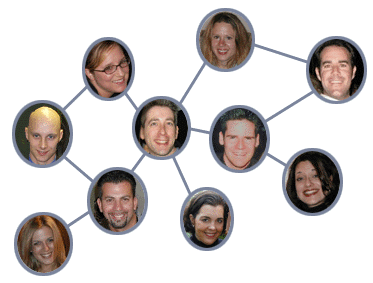
Finally, we have a drawing of the proposed ARPANET in the late 1960s:
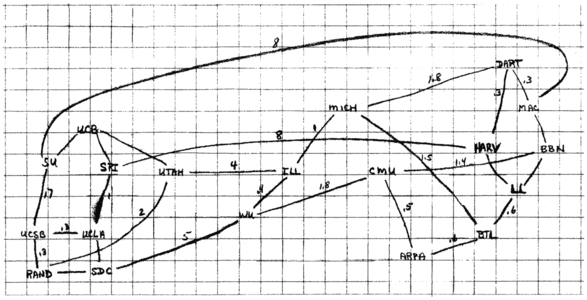
There are a lot of questions we can ask about these types of relationships: What is a route between Suitland and Foggy Bottom (I think we can agree those are the two silliest Metro station names)? Does Jack know Jill? Does Jack know somebody who knows Jill?
All of these graphs are undirected, which means that the pathways run both ways: people are friends with each other, and a path between New Carrollton and Landover implies a path between Landover and New Carollton. But, they don’t have to be. Google represents the internet using a directed graph: a directed edge between two pages P1 and P2 exists if P1 links to P2.
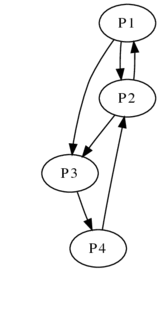
Simplistically speaking, Google calculates the probability of ending up at a certain page given random clicks (equating to a random walk around the graph). The higher the probability that you end up on a page, the more influential and important it is.
So those two graphs are directed, but they are all still unweighted. Consider this graph:
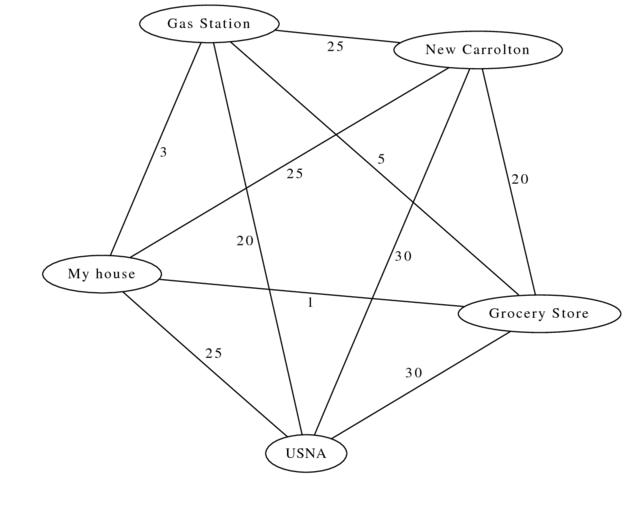
In this graph, the edges are weighted by the amount of time it takes to get between different places. If I’m home, and need to go to work, the grocery store, and the gas station, what’s the fastest path?
So graph algorithms are really, really important, and we’ll be spending some time on them.
2 Graph Terminology
Node or vertex: A single, labeled spot on the graph (usually drawn as the node label in a circle or oval).
Edge: A connection between two nodes.
Graph: A collection of nodes and edges between those nodes.
Directed graph: Graph in which all edges do have an arrow indicating a particular direction.
Note, this doesn’t mean everything is only one-way; you can have two directed edges between a pair of nodes, one going each way.
Undirected Graph: Graph in which edges have no arrows or direction. Equivalently, can be thought of as a directed graph where every edge goes both ways.
Weighted graph: Every edge is associated with a numerical value.
Adjacent Vertices: Vertices (i.e., nodes) which are connected by an edge are called adjacent.
Degree: The number of edges connected to a vertex. If there are \(m\) edges in a graph, \(\sum_{v\in G} deg(v) = 2m\) (because each edge is connected to two vertices).
In degree and out degree: Number of directed edges coming into or going out of a vertex respectively.
Path: A sequence of vertices, such that each vertex is adjacent to the vertices preceding and succeeding it.
Two vertices are connected if there is a path that goes from one to the other.
There are also some variable names that are used pretty consistently, so let’s write them down here so we can all get on the same page:
\(V\): the set of all vertices
\(E\): the set of all edges
\(u, v\): usually the names of vertices
\(w\): usually the weight of some edge
\(G\): a complete graph, which is just a pair \((V,E)\).
\(n\): the number of vertices in the graph
\(m\): the number of edges in the graph
3 Graph ADT
The precise interface for dealing with a graph will depend slightly on whether it’s weighted or unweighted, and directed or undirected. We’ll describe an abstract data type for a directed and weighted graph; it might be just a little bit simpler if the graph doesn’t have directions or weights.
getVertices(): returns a list of all vertex labels in the graphgetEdge(u, v): returns the weight of the edge fromutov, ornullif there is no such edgeneighbors(u): returns a list of edges that start from the vertex labeleduaddVertex(u): adds a new vertex labeledu, if it doesn’t already exist. The new vertex will initially have no neighboring edges.putEdge(u, v, w): adds a new edge fromutovwith weightw, or changes the existing weight if the edge already exists.
4 Representing a Graph
Now we have a graph ADT, so what will the data structures be to implement that ADT? Well there are actually two things to represent here: the vertices, and the edges.
To represent the vertices, we need a Map that will associate any vertex name (probably a string, but maybe something else) to that vertex’s representation in the data structure. So the keys in this map will be vertex labels. The best choices would be a balanced search tree or a hashtable.
4.1 Adjacency lists
What will the “representation in the data structure” for each vertex be? Well that depends on how we are storing the edges. First, you can use adjacency lists. In an adjacency list representation, each vertex is represented by a list of edges going out from that vertex. (And each edge object should contain the destination vertex as well as the weight where applicable.) In the case of an adjacency list, your vertex map would have labels as the keys and lists of edges as the values.
For example, here is a weighted, directed graph:
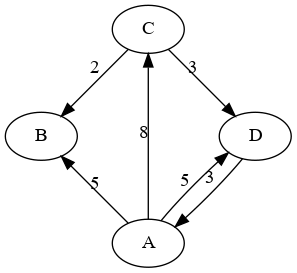
The adjacency lists representation of this graph would be a map from strings to lists of edges, something like this:
A: [ (B,5) (C,8) (D,5) ]
B: [ ]
C: [ (B,2) (D,3) ]
D: [ (A,3) ]Notice that there is one list for each vertex, and the size of each list is the out-degree of that vertex. In an unweighted graph, we would just have lists of adjacent vertex labels (no weights), and in an undirected graph, each edge would appear in both adjacent nodes’ lists. It is also important to point out that, although they are in the example above for clarity, the map and lists are not necessarily sorted.
4.2 Adjacency matrix
The second option to represent a graph is an adjacency matrix. For this, if we have \(n\) vertices, we construct a \(n \times n\) matrix using a double array. Each vertex is simply represented by an index from 0 to \(n-1\), so that every vertex has a row and a column dedicated to it. If location \((i,j)\) is null, no edge connects \(V_i\) and \(V_j\). If an edge does connect those two vertices, location \((i,j)\) will contain the weight of that edge. (In an unweighted graph, we would store boolean true/false values at each matrix location).
Here is how the same example graph above would look as an adjacency
matrix. Notice there are two parts: one map from vertex names to
indices, and then the 2D array for the weights of all edges.
In this diagram, x is used to indicate a null value.
{A:0, B:1, C:2, D:3}
0 1 2 3
0 [ x 5 8 5 ]
1 [ x x x x ]
2 [ x 2 x 3 ]
3 [ 3 x x x ]Usually, for convenience, we don’t draw the vertex names map separately, but just draw the vertex labels at the row and column labels on the matrix itself.
Also, depending on the actual application, it’s common to not use nulls here but rather special values, especially when the edge weights represent distance in some way:
Every vertex going to itself along the “main diagonal” (as in indices
(0,0),(1,1),(2,2), etc.) in the matrix has weight 0.This represents the idea that the distance of a node to itself is 0.
Every edge that doesn’t exist is represented with weight infinity, usually written \(\infty\).
This represents the idea that the distance between two nodes that are not connected is infinitely large.
Note that, in most programming languages, you can represent positive or negative infinity as a floating-point value. In Java, you can get it with
Double.POSITIVE_INFINITYfor example.
4.3 Comparing the representations
There must be pros and cons to each of these. Let’s call the number of vertices \(n\), and the number of edges \(m\). Most of the asymptotic costs we can think about with graphs are in terms of both \(n\) and \(m\).
But these variables are not completely independent of each other! If you think about it, the number of edges can never be more than \(n(n-1)\) in a directed graph, which is \(O(n^2)\). Sometimes we can simplify a big-O with this fact; for example \(O(n^3 + m\log n)\) could be simplified to just \(O(n^3)\).
Memory usage: For an adjacency list, there are \(n\) independent lists, and each one has at most \(n-1\) nodes. So the total size is \(O(n^2)\). But that’s actually an over-estimate! The total size of all lists equals the number of edges, \(m\). So actually the total size of the adjacency lists representation is \(O(m+n)\).
An adjacency matrix, on the other hand, has an entry for each possible edge, whether it exists or not, meaning \(O(n^2)\) space, which is not an over-estimate.
So asymptotically, the adjacency matrix will use the same or possibly more memory than adjacency lists. But in practice, when the graph has many edges (i.e., \(m\) is close to \(n^2\)), the adjacency matrix may be smaller due to not having to explicitly write down the labels of each edge.
getVertices(). This is a simple traversal through the map, which should always be \(O(n)\) time.getEdge(u,v), i.e., testing whether a certain edge exists and if so finding its weight. First we have to look upuandvin the map. Then, in an adjacency matrix the edge is just an entry in the double array, \(O(1)\) time to look it up. In an adjacency list, the cost depends on how many neighborsuhas, so it’s \(O(\deg u)\), which could be as much as \(O(n)\).neighbors(u), i.e., returning a list of all vertices incident to a certain vertex: For an adjacency list, this list already exists, you merely have to return a pointer to it. So, this is just the time it takes to lookup that vertex in the map. For an adjacency matrix, you need to iterate over that row of the matrix and create the list for all non-null (or non-infinite) entriex, which costs \(O(n)\).addVertex(u):, i.e., inserting a vertex: For an adjacency list, this merely requires adding a new vertex to the list of vertices, which is just the cost of theputoperation in the map. For an adjacency matrix, you have to add a new row and column to the matrix, which is \(O(n)\) amortized time. (Because sometimes the underlying arrays will have to be doubled in size, costing \(O(n^2)\) in the worst case.)putEdge(u,v,w), i.e., inserting or updating an edge: For both, this will have the same cost ofgetEdge, since actually inserting or updating the list after searching is \(O(1)\), and updating a single matrix entry after looking up the edge labels is also \(O(1)\).
We can summarize all this as follows. As always, we emphasize that you should not feel the need to memorize this table. Yes, some portion of this will be on your exam. But memorizing it is not a good use of your time. Instead, you should be able to reason through each running time as we did in the discussion above. If you understand what a graph is and how our basic data structures (arrays, linked lists, balanced trees, hashtables) work, then you should be able to fill this in without any rote memorization!
| getVertices | getEdge | neighbors | addVertex | putEdge | |
|---|---|---|---|---|---|
| Adj list w/ balanced tree | \(O(n)\) | \(O(n)\) | \(O(\log n)\) | \(O(\log n)\) | \(O(n)\) |
| Adj list w/ hashtable | \(O(n)\) | \(O(n)\) | \(O(1)\) avg | \(O(1)\) avg | \(O(n)\) |
| Adj matrix w/ balanced tree | \(O(n)\) | \(O(\log n)\) | \(O(n)\) | \(O(n)\) amort | \(O(\log n)\) |
| Adj matrix w/ hashtable | \(O(n)\) | \(O(1)\) avg | \(O(n)\) | \(O(n)\) amort | \(O(1)\) avg |
5 Depth First Search of a Graph
One of the things we want to do with just about any data structure is traverse it, meaning to iterate over all the things in that data structure in some kind of order. We talked about 4 different ways to traverse trees; what changes when we want to traverse a graph instead?
The first thing that changes is the name: traversing a graph is called graph search as it is “exploring” the graph in some way. And while tree traversals always start from the root, a graph search can start from anywhere. So a graph search always has a particular starting vertex; it might have an ending (or “destination”) vertex too, like if we are searching for a particular path through the graph.
If we took the usual pre-order traversal algorithm from trees and applied it to
graphs, it would work except for one small problem: any cycle in the graph
would result in an infinite loop in the algorithm! So we have to add one more
thing, a set visited which stores which nodes have been traversed already.
The pseudocode for this search might look something like this:
DFS(G, start):
visited = new Set of vertices
DFS_Helper(G, start, visited)
DFS_Helper(G, uu, visited):
if ! visited.contains(uu):
visited.add(uu)
print(uu.data)
for each node vv in G.neighbors(uu):
DFS_Helper(G, vv, visited)
This is called a Depth-First-Search, or DFS for short, because of the way it explores the graph; it sort of “goes deep” before it “goes wide”.
That’s a mighty fine recursive algorithm, but of course we could write it iteratively too if we wanted. The main difference is that we will need a stack to store the nodes that are currently being explored. This essentially takes the place of the function call stack that the recursion uses implicitly.
DFS(G, start):
visited = new Set of vertices
fringe = new Stack of vertices
fringe.push(start)
while fringe is not empty:
uu = fringe.pop()
if ! visited.contains(uu):
visited.add(uu)
print(uu.data)
for each node vv in G.neighbors(uu):
fringe.push(vv)
DFS is great when we are trying to just traverse the graph and we don’t
care about the order, or if we are trying to learn something about
cycles or ordering relationships in a directed graph. For example, if
you had a graph and wanted to know whether or not it contained any
cycles, you could do a DFS starting from each vertex, and check if each
child node vv is already in the fringe before adding it on the last
line. If we come across a node in the neighbors of uu that is already
in the stack, then we’ve found a cycle in the graph!
6 Breadth First Search of a Graph
Let’s go crazy and replace the stack from DFS with a queue instead. What happens is that we get something more like a level-order traversal of a tree, where the nodes closest to the starting vertex are explored first. This is called a breadth-first search (BFS), and it can be used to find the shortest path between two vertices in an unweighted graph.
Here’s pseudocode for BFS, where we also added in a map of “parents” that keeps track of the shortest paths, for purposes of backtracking and recovering that actual path later.
Note a subtle difference between the visited set of DFS and the
discovered set used here: A node is considered “discovered” as soon as
it is added to the fringe, and “visited” after it gets removed from
the fringe. Using a discovered set is a little more memory-efficient
because the fringe gets fewer things added to it, but it’s only possible
with BFS. (Since a queue is FIFO, there is no need to add anything to it
a second time. Not so with a stack!)
BFS(G, start):
discovered = new Set of vertices
fringe = new Queue of vertices
parents = new Map from vertices to vertices
fringe.enqueue(start)
discovered.add(start)
while fringe is not empty:
uu = fringe.dequeue()
print(uu.data)
for each node vv in G.neighbors(uu):
if ! discovered.contains(vv):
fringe.enqueue(vv)
discovered.add(vv)
parents.put(vv, uu)Note that if there was a particular destination vertex we were searching
for, the BFS could be terminated as soon as that vertex is “discovered”
in the nested if statement.
Recovering the path is just a matter of following the vertices in
parent until you get back to the start.
Here’s an example of this running, using a “word ladder” to demonstrate. Here, each vertex is an English word, and if two words are exactly one letter apart (like “cape” and “cane”), then an edge is drawn between then. We’re searching for a path from “rust” to “belt”.
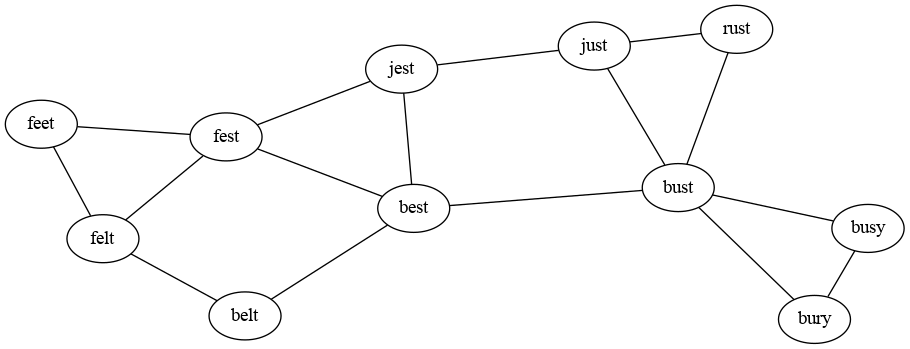
Initially we start with “rust” in the discovered set and the fringe.
discovered = { rust }
fringe = [ rust ]
parents = { }Now we dequeue “rust” and add its undiscovered neighbors to the
discovered set and the back of the fringe. We also set both of those
neighbors’ “parent” to rust.
discovered = { rust just bust }
fringe = [ just bust ]
parents = { just: rust
bust: rust
}The next word to be dequeued is “just”. This has three neighbors in the
graph, but only “jest” is not yet in the discovered set, so that is the
only new entry added to the fringe and to the parents map.
discovered = { rust just bust jest }
fringe = [ bust jest ]
parents = { just: rust
bust: rust
jest: just
}Next we dequeue “bust”, which has five neighbors in the graph, two of
which have already been discovered. So we add three new words to the
three data structures.
discovered = { rust just bust jest best busy bury}
fringe = [ jest best busy bury ]
parents = { just: rust
bust: rust
jest: just
best: bust
busy: bust
bury: bust
}“Jest” is next, discovering one new word:
discovered = { rust just bust jest best busy bury fest }
fringe = [ best busy bury fest ]
parents = { just: rust
bust: rust
jest: just
best: bust
busy: bust
bury: bust
fest: jest
}Finally, when we dequeue “best”, we will discover our target word “belt” and can stop the search with the following data structures:
discovered = { rust just bust jest best busy bury fest belt }
fringe = [ busy bury fest belt ]
parents = { just: rust
bust: rust
jest: just
best: bust
busy: bust
bury: bust
fest: jest
belt: best
}Finally, from the parents map, we can follow the links backwards to
discover the path back to the starting point: belt–best–bust–rust.
Note that the FIFO order of the queue is very important here! If we had a stack instead (as in DFS), we would still find “belt” from “rust”, but might take a much longer path like rust–just–jest–fest–feet–felt–belt.
So, what does BFS guarantee us? Well, first, the only thing that causes this to stop, is we either find v, or we run out of connected, unvisited vertices. So, if a path exists (if v is connected to u), then we are guaranteed to find it. Additionally, because all vertices that can be reached in \(k\) steps are added before any vertices that can only be reached in \(k+1\) steps are added, the path we find will be the shortest such path that exists. Cool!
Credit Randall Munroe, XKCD.com
It’s worth noting that the BFS and DFS algorithms don’t change if we’re looking at a directed graph.
7 Runtime of graph search
Analyzing these algorithms requires us to understand a lot of data structures!
- The graph has a size-\(n\) map of vertex names (balanced tree or hashtable)
- The graph also contains the edge data in adjacency lists or an adjacency matrix.
- The
fringeis a stack or a queue of size at most \(m\) or \(n\) respectively (depending on DFS or BFS) - The
visitedordiscoveredsets have size at most \(n\) and are searched usingcontains()at most \(m\) times; they can also be either balanced trees or hashtables.
Using adjacency lists and balanced trees, and linked lists for the stacks or queues, the running time of both BFS and DFS is \(O((n+m)\log n)\). Using hashtables for the maps and sets instead reduces this to \(O(m+n)\) on average. That’s very fast considering the graph has \(n\) vertices and \(m\) edges.
Using an adjacency matrix for the graph, the cost to call neighbors()
\(n\) times already gives us \(O(n^2)\) total; then we should use hash
tables for the visited or discovered sets so that the total cost is
\(O(n^2)\) average.
But there is a small optimization to do here: we only really need
to look up the labels for the starting and ending vertices in the map,
and the rest of the time we can use the indices directly. So the
visited or discovered sets can just be implemented with a size-\(n\)
array of booleans, and we get \(O(n^2)\) worst-case time for both DFS
and BFS.
We emphasize again: it is good to know these costs of DFS and BFS for the various choices of data structures, but even more important that you are able to derive these running times from the pseudocode above.